
home > research > SFB 902
Structure and conformational flexibility of RNA studied by EPR
spectroscopy
(project within SFB 902: Molecular Principles of RNA-based Regulation)
This project aims to investigate structural and conformational flexibility aspects of RNA and RNA-protein complexes by continuous-wave (cw) EPR and Pulsed Electron-Electron Double Resonance (PELDOR) spectroscopy performed at several magnetic field values (from 0.1 to 6.4 T). Spin-labeled RNA is used to explore structure, conformational flexibility and dynamics of tertiary folded RNA motives as well as the structural transitions induced by binding to proteins, metal ions or ligands. Our preliminary work on nucleic acid molecules is summarized in the following book chapters. [1]
Quantitative characterization of small RNA motives
We investigate structure and conformational flexibility of several small RNA motives with different three-dimensional architecture and compare
our multi-frequency, multi-field PELDOR analysis with data from NMR, X-ray, FRET and MD studies on the same systems to evaluate the potential of
the PELDOR method in this respect. The question to be answered is, whether it is possible to get a unique solution of the conformational flexibility
of such RNA motives. Furthermore we ask if the conformational flexibility determined by PELDOR at low temperatures can be related to the conformational
dynamics occur at physiological conditions.
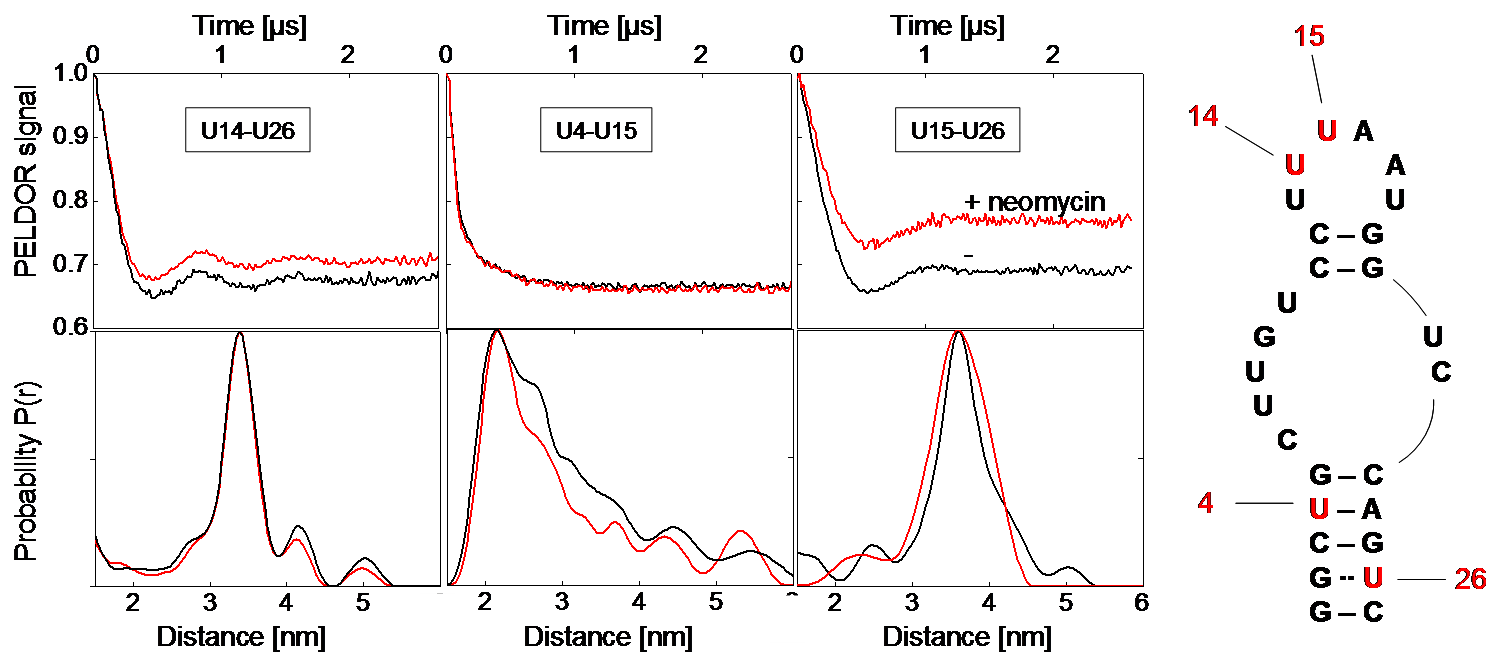
Figure 1: Left: Background-corrected PELDOR time traces and corresponding distance distributions of the three double-labeled neomycin-responsive riboswitch samples in presence (red) and absence of neomycin (black). Right: Secondary structure of the neomycin-responsive riboswitch with positions of spin-labeled nucleotides. [2]
Evaluation of spin-labeling strategies for long RNA molecules and in-cell applications
The development of spin-labeling techniques for RNA molecules is one of our main issues. The long-term goal is to explore the possibility of applying PELDOR under physiological conditions (room temperature, in-cell) to large RNAs or RNA/protein complexes. This project is done in close collaboration with the research group of Prof. Göbel, who develops and synthesizes new type of spin labels for this purpose and use ligation techniques to incorporate them into larger RNA molecules. In collaboration with Prof. Sigurdsson (University Island) we investigate in the rigid spin label Ç incorporated in DNA or RNA for different application e. g. in-cell.
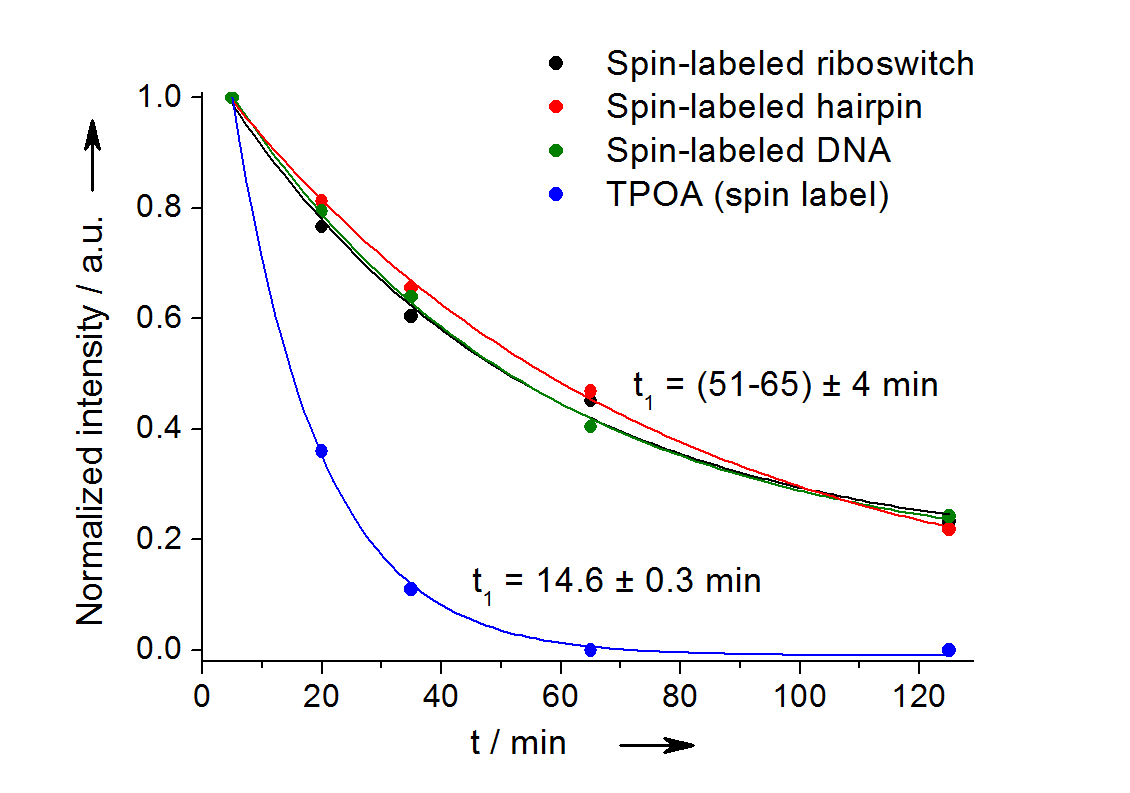
Fig 2: In-cell reduction kinetic curves for the spin label nitroxide attached to a 27-mer neomycin-sensing riboswitch (black), a 14-mer RNA hairpin (red), a 12-bp duplex DNA (green) and for the free spin label (blue). [3]
Investigation of RNA-protein complexes and RNA-folding dynamics
We study structural aspects of small RNAs bound to ribosome assembly factors and obtain long-range distance restraints of freeze-quench trapped folding intermediates of RNA riboswitches. In collaboration with the research group of Prof. Tampé, we work on studying the conformational changes of the ABCE1 release protein within the ribosome recycling.
Development of new strategies for the analysis of orientation selective PELDOR data
If rigid spin labels are incorporated into the DNA or RNA, the relative orientations between a pair of such spin labels can be extracted from orientation selective PELDOR experiments in addition to provide their distance. This allows the determination of both the structure and the mobility of the nucleic acid.[4] We are developing new strategies to obtain orientation information from this PELDOR data and thereby geometrical restraints for structure determination. To improve the uniqueness of the solution for the overall structure of the nucleic acid from only a few doubly spin-labeled mutants, we will perform orientation selective PELDOR experiments at different microwave frequencies and magnetic fields.
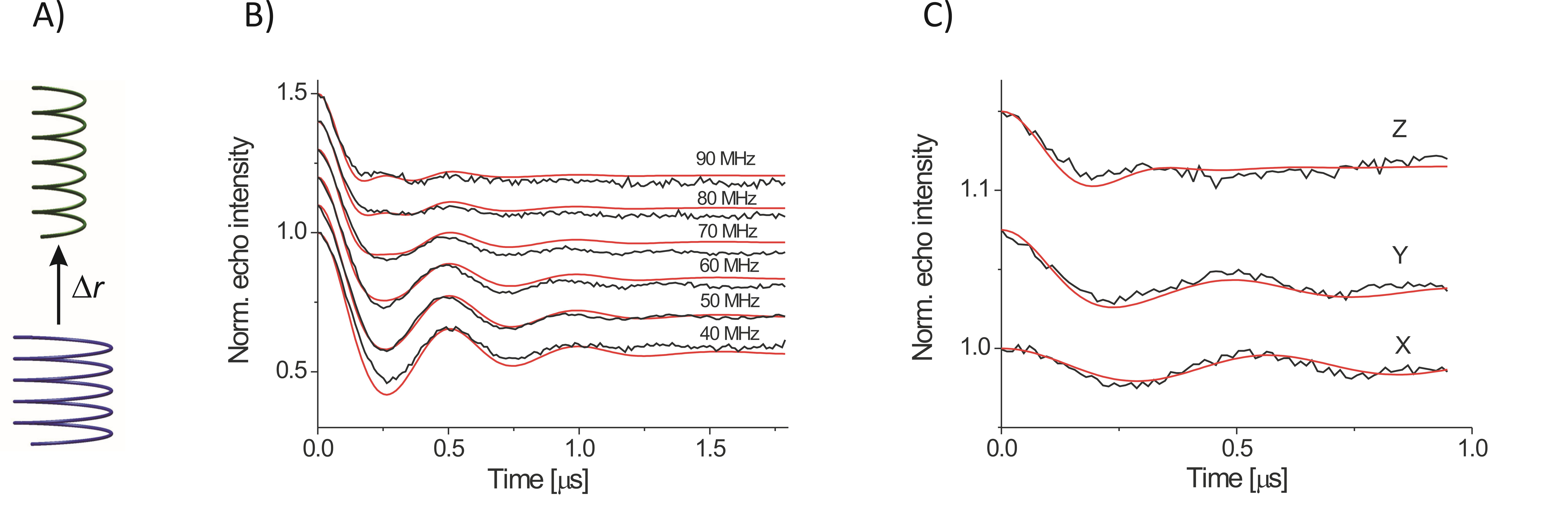
Fig. 3: A. Twist-stretch dynamic model of double-stranded DNA. B. and C. Comparison of the X- and G-band experimental orientation selective PELDOR data with simulations based on this model. [1b, 4b]
References:
| [1] | a) I. Krstic, B. Endeward, D. Margraf, A. Marko, T. F. Prisner, in Epr Spectroscopy: Applications in Chemistry and Biology, Vol. 321 (Eds.: M. Drescher, G. Jeschke), Springer-Verlag Berlin, Berlin," 2012, 159-198; b) I. Krstic, A. Marko, C. M. Grytz, B. Endeward, T. F. Prisner, in RNA Structure and Folding: Biophysical Techniques and Prediction Methods (Eds.: D. Klostermeier, C. Hammann), De Gruyter, 2013, 261-286. |
| [2] | I. Krstic, O. Frolow, D. Sezer, B. Endeward, J. E. Weigand, B. Suess, J. W. Engels, T. F. Prisner, J. Am. Chem. Soc. 2010, 132, 1454-1455. |
| [3] | I. Krstic, R. Hansel, O. Romainczyk, J. W. Engels, V. Dotsch, T. F. Prisner, Angew. Chem.-Int. Edit. 2011, 50, 5070-5074. |
| [4] | a) O. Schiemann, P. Cekan, D. Margraf, T. F. Prisner, S. Th. Sigurdsson, Angew. Chem.-Int. Edit. 2009, 48, 3292-3295; b) A. Marko, V. Denysenkov, D. Margraft, P. Cekan, O. Schiemann, S. Th. Sigurdsson, T. F. Prisner, J. Am. Chem. Soc. 2011, 133, 13375-13379. |
Collaborators within the SFB 902:
A1 Schwalbe (riboswitch folding, model-RNAs)
A2 Suess (aptamers, in-cell)
A4 Göbel (spin label developments: int. al. photo-protected spin labels)
B3 Göringer (editosome/RNA)
B7 Tampé (ABCE1)
B10 Wöhnert (Nep1/RNA, methods)
External Collaborators:
Snorri Th. Sigurdsson (University Island, Reykjavik, Island)
Claudia Höbartner (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany)
Michael Sattler (Technische Universität München, Munich, Germany)
Theresa Carlomagno (EMBL, Heidelberg, Germany)
| Last modified: April 08, 2014 | Impressum / About / Legal Notice | Datenschutzerklärung |
Prisner LOGS
Webmail
Internal
|